Neuroanatomy of DBS-OCD using advanced imaging
The goal of this project (funded R01 project titled “Patient-specific, Effective, and Rational Functional Connectivity Targeting for DBS in OCD”) is a retrospective investigation of imaging-based targeting for Deep Brain Stimulation (DBS) for severe obsessive-compulsive disorder (OCD). We use anatomic and functional connectivity analyses, plus electrical modeling, to identify white and grey matter zones that must be captured by the electrical stimulation to yield clinical response. [https://reporter.nih.gov/search/QhVboFoXdUe_XiAriH9B5w/project-details/9750115]
Multimodal imaging in Gambling Disorder
In this R21 project titled “Modulating Inhibitory Control Networks in Gambling Disorder with Theta Burst Stimulation”, we are implementing multimodal structural magnetic resonance imaging (MRI) and functional MRI methods of analysis in a population of patients affected by gambling disorder undergoing neuromodulation for cognitive control in order to further our understanding in these areas of inquiry. [https://reporter.nih.gov/search/QhVboFoXdUe_XiAriH9B5w/project-details/9566150]
Comparative neuroanatomy of white matter fiber tracts using dMRI tractography
Diffusion MRI derived white matter tractography allows tracing of the neural fiber connections in the human and primate brain. Ground truth data about the white matter topology can, however, only be obtained from tracer studies in non-human primates. Our current projects involve developing a comparative (homologous) white matter atlas of long range, medium range and the superficial white matter for human and non-human primates using dMRI tractography (see Figs 1 and 2). Data from in-vivo and ex-vivo high resolution human and monkey brains are used to trace fibers using our award-winning (Fillard et al., 2010) Unscented Kalman Filter based multi-tensor tractography algorithm (Malcolm et al., 2010, Reddy et. al., 2016). Our white matter atlas was created using the clustering algorithm developed by our collaborators Drs. Lauren O’Donnell and Fan Zhang (Zhang et.al., 2018).
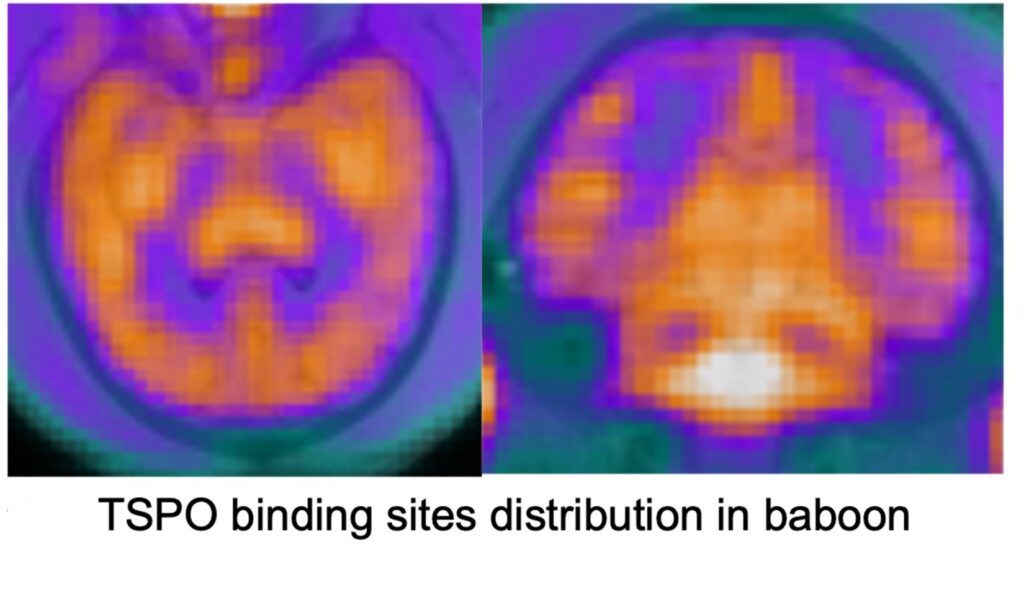
Imaging neuroinflammation in aging non-human primates
Chronic neuroinflammation and subsequent myelin degeneration are believed to play crucial roles in cognitive aging. At the CMA, we use animal models to develop and validate various neuroimaging biomarkers sensitive to those biological processes. For example, we have demonstrated the relationship between activated microglia and cognitive impairment in rhesus monkeys (Shobin et al., 2017), we have investigated molecular pathways of white matter vulnerability in aging monkeys (Robinson et al., 2018), and we have shown trajectories of white matter in vivo diffusion MRI biomarkers of neuroinflammation and myelin breakdown (using free water imaging) access the life span trajectory of rhesus monkeys (Kubicki et al., 2019). Our current projects involve imaging and validation of multimodal MR methods sensitive to neuroinflammation and myelin degeneration in rhesus monkeys. Those methods involve diffusion free water imaging, arterial spin label and positron emission tomography(Histone deacetylases (HDAC), translocator protein (TSPO)).
Tool development and data analysis in schizophrenia
Neuroimaging plays a crucial role in better understanding schizophrenia pathology. While various methods over the years have demonstrated neural abnormalities in this disorder, small sample sizes as well as low biological specificity of existing methods continuously hamper the research. To overcome small sample size limitations, we have developed and validated diffusion imaging harmonization procedure (Mirzaalian et al., 2018; Cetin-Karayumak et al., 2019) that allows for a diffusion data harmonization at the signal level. We have applied this harmonization tool to 13 schizophrenia datasets we have gathered (1200 cases), and investigated diffusion changes along the schizophrenia time course (Cetin-Karayumak et al., 2020), as well as clinical and cognitive correlates of imaging changes in this large sample (Seitz-Holland et al., 2021). To increase the biological specificity of neuroimaging measures, we have developed a free water imaging technique, which demonstrates the sensitivity of free water (FW) measures to acute brain changes in first episode psychosis (Pasternak et al., 2012; Lyall et al., 2018), and demonstrated increased FW in a rat model of immunological response (Di Biase et al., 2020), as well as relationship between FW and peripheral proinflammatory cytokines in schizophrenia (Di Biase et al., 2021).
Transcranial magnetic stimulation in Depression
Transcranial magnetic stimulation (TMS) is a non-invasive neuromodulation technique that provides a new paradigm for treating brain diseases including major depressive disorder (MDD) and obsessive compulsive disorder (OCD). TMS works by placing a coil of wires enclosed in a plastic case near the surface of the scalp to induce electrical current changes underlying brain tissue. Since human subjects have significant inter-subject variabilities in brain anatomy and function, personalized targeting of TMS based on subject-specific brain connectivity has the potential to improve treatment efficacy. Our team is collaborating with the neuropsychiatry modulation lab at MGH led by Dr. Joan Camprodon to investigate subject-specific white-matter connections obtained by diffusion MRI that are relevant to TMS treatment response. The microstructural changes in white matter pathways induced by TMS treatment can provide insights for understanding the mechanism of action leading to a potentially more effective targeting approach. To improve targeting accuracy, our team has been developing new computational tools that use a deep-learning approach to predict the electric field distribution in brain tissue. Rapid electric field prediction is useful to improve the placement of TMS coils to optimize the stimulation of brain target areas or white matter pathways, thus enhancing treatment efficacy.
[https://reporter.nih.gov/search/xXvt0xXe00qqEW5EGRJ52w/project-details/10195450]
Methods and Tools:
Tractography and white matter atlas:
Diffusion MRI allows the determination of fiber orientations of white matter at the millimeter scale. While several tractography algorithms have been described in the literature, most of these estimate the fiber orientation distribution functions (ODF) independently at each voxel and trace the connections from a given seed point in the white matter to its termination in gray matter. However, the estimated ODFs are typically afflicted with noise and hence independent estimation of the ODF at each voxel can lead to very noisy tracts. Our Unscented Kalman Filter (UKF) based tractography algorithm (Malcolm et al., 2010, Reddy et. al., 2016) however, uses the correlation in the motion of water molecules along the fiber bundle to simultaneously trace the fibers while estimating the parameters of the underlying model (e.g. multi-tensor, free-water, NODDI, diffusion propagator, etc.). This ensures regularized and robust estimation of not only the tracts but also the microstructural parameters. Our algorithm also provides a measure of confidence in the obtained fit at each location along the tract. Our algorithm was one of the winners of the Fiber Cup community competition held during MICCAI 2010 (Fillard et al., 2010).
We have developed the most comprehensive human white matter atlas for long and medium range connections in human brains based on connectivity information from non-human primate tracer studies. This atlas (Zhang et.al., 2018) has been shown to consistently extract tracts across the life span (neonate to 80+ years) as well as in the presence of large tumors.
dMRI and sMRI processing pipeline
We use the latest state-of-the-art methods for data pre-processing developed by our collaborators at the Psychiatry Neuroimaging Laboratory (PNL) ; Brigham and Women’s Hospital. A detailed note about the pre-processing steps for diffusion and structural MRI are noted on the following Github page: https://github.com/pnlbwh/
Our workflow includes not only motion, eddy and geometric distortion correction, but also algorithms for data QC as well as a Luigi pipeline for batch processing of a study to ensure consistent processing across all subjects.
Advanced fMRI analysis
Diverse functions of the human brain arise from neuronal interactions between different regions this form the networks of the human brain. Diffusion MRI (dMRI) and functional MRI (fMRI) are neuroimaging techniques that probe the structural substrates of brain networks and brain activities’, respectively. Integrating information from the two techniques can provide insights for understanding the structure-function relations of brain networks that underlie mechanisms of mental disorders. Our team has developed a set of computational tools based on control and signal processing theories for joint analysis of dMRI and fMRI data. The multimodal measures based on the joint analysis can provide novel markers for brain diseases such as schizophrenia and depression (Ning et al., 2012, Ning et al., 2017, Ning et al., 2017)
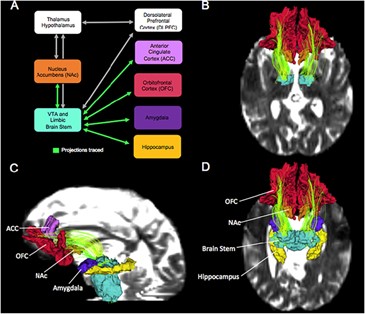
Neuroanatomy of alcohol use disorders
The goal of this project is to use multimodal imaging in conjunction with behavioral tests to identify changes in the structure and function of subcortical circuits in abstinent alcoholics. Subcortical brain circuits connecting structures such as the nucleus accumbens, amygdala, hypothalamus and brainstem are critical for understanding substance use disorders such as alcohol use disorders, but are difficult to resolve using standard methods. The goal of this project is to examine the structure and function of subcortical circuits using high resolution imaging to determine how the structure, function and connectivity of these circuits change in male and female abstinent alcoholics in conjunction with related behaviors.
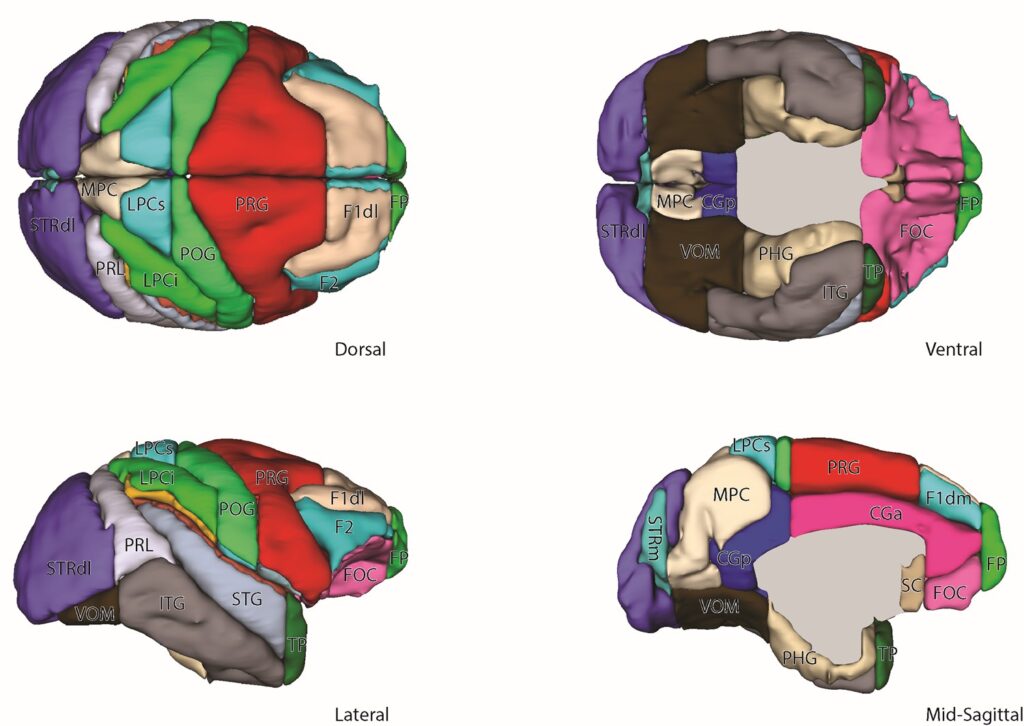
Comparative neuroanatomy using the Harvard-Oxford Atlas (HOA) system
The HOA is a widely used method to parcellate brain structures in the human as visualized using MRI. The goal of this project is to use homological relationships to extend the HOA methodology and generate a comparative parcellation framework for the macaque monkey brain.
Method/tool development and clinical application
The Center for Morphometric Analysis has been at the forefront of MRI-based neuroanatomy since the inception of MRI methods and has developed many of the fundamental tools and procedures for manual and semiautomated morphometric analysis of brain structure. These procedures and tools were originally part of the CardViews software package at the CMA. We are currently developing similar and novel tools to support manual editing of brain MRI data in the 3D Slicer environment.
Manual for subcortical brain segmentation
The Center for Morphometric Analysis has always been at the forefront of manual segmentation of brain structures. We are currently developing and perfecting tools and protocols for segmentation and parcellation of high resolution structural MRIs.